5-HMR-16 Virtual Coupling
The term "virtual coupling" refers to an NMR phenomenon in which apparently first-order multiplets contain false coupling information. In extreme cases, protons that are not actually coupled will show splitting. More commonly, the magnitude of coupling constants obtained by first-order analysis is incorrect. All virtual coupling effects arise when protons, well isolated from other protons in chemical shift, are coupled to a group of other protons which are strongly coupled to each other. By strongly coupled we mean that these protons are both close in chemical shift and coupled to each other with J > Δν.
On the following pages are examples which illustrate the virtual coupling phenomenon in several different systems. One simple way to summarize these effects is as follows: when two or more protons are strongly coupled, then any protons coupled to them will give multiplets with false coupling information and/or unexpectedly complex multiplet structure. Furthermore, the strongly coupled protons themselves will contain misleading multiplet structure. Virtual coupling effects are also frequently encountered in heteronuclear magnetic equivalence NMR situations, e.g. in transition metal phosphine complexes (see Sect. 7-MULTI-2)
The simplest systems to show virtual coupling effects are ABX patterns, where the X-multiplet will give incorrect values for JAX and JBX when it is analyzed as a doublet of doublets if νAB is similar to or smaller than JAB. What is particularly insidious about this effect is that analysis of the AB part by a first-order method (treatment of each part as a distorted doublet of doublets) will give the same incorrect values for JAX and JBX.
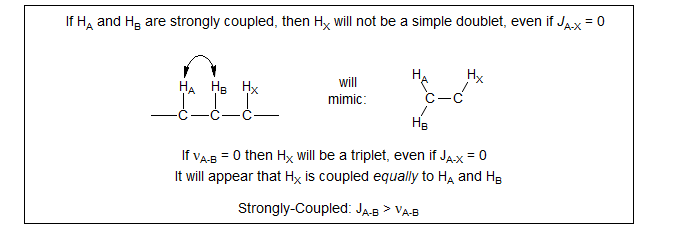
A typical example of virtual coupling is provided by the epoxide below. HA and HB are more or less first order when the spectrum is taken in CDCl3 because HA and HB have a significant chemical shift (although typical second order effects are starting to appear). However, in acetrone-d6, HA and HB are essentially superimposed, and HC appears as a triplet, as if HA and HB were equally coupled to HC, leading to a very different structure assignment. The small coupling visible for HB and HC in CDCl3 is probably not entirely real - it is the beginning of the "virtual coupling" effect which eventually leads to a triplet for HC
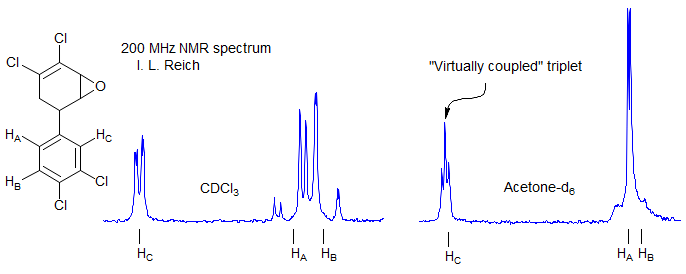
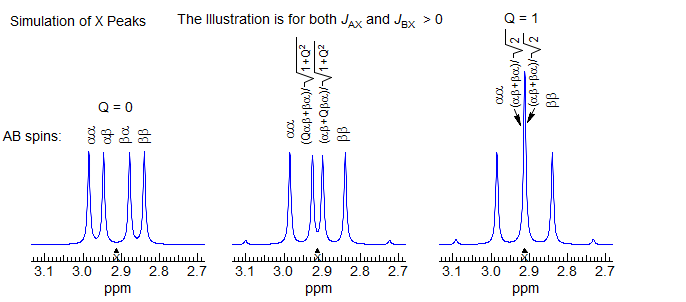
These effects can be understood by examining the behavior of the X part of an ABX system as ΔνAB becomes small. In this simulation, JAX is set to 0; nevertheless when ΔνAB < 15 Hz the X part shows clear indications of coupling to HA, as does the AB part, i.e. virtual coupling.
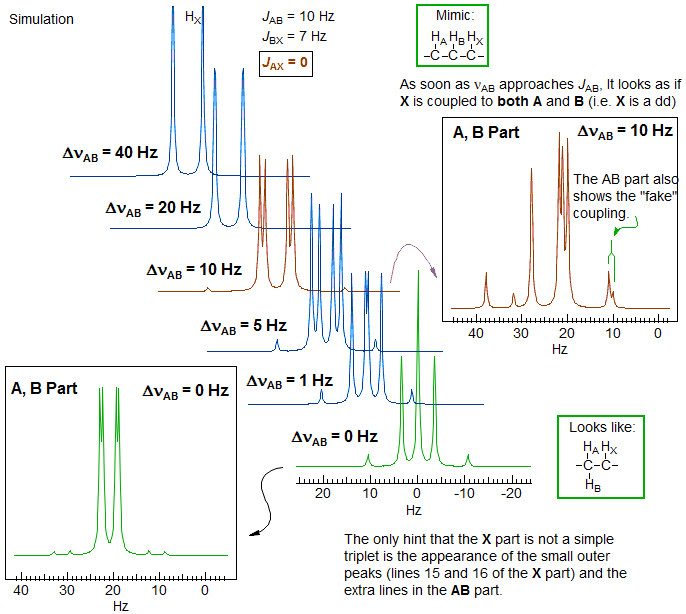
Virtual Coupling in X of an ABX system
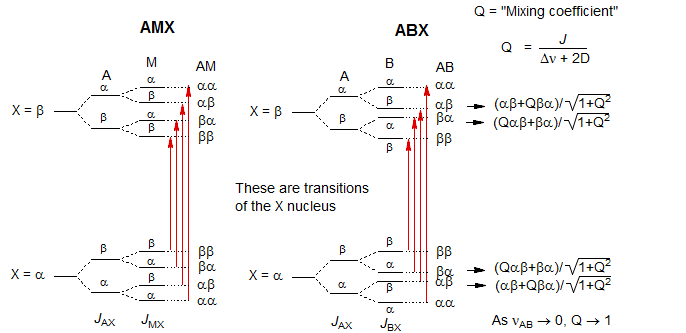
As the chemical shift between HA and HB becomes smaller, the αβ state begins to mix with the βα state, and the βα state mixes with the αβ (the mixing coefficient is Q). This results in their energies becoming closer together, and eventually, when νAB = 0, Q becomes 1, and the two energy levels consist of equal parts αβ and βα, and their energies are identical (the X part becomes a triplet).
"Virtual Coupling" Effects
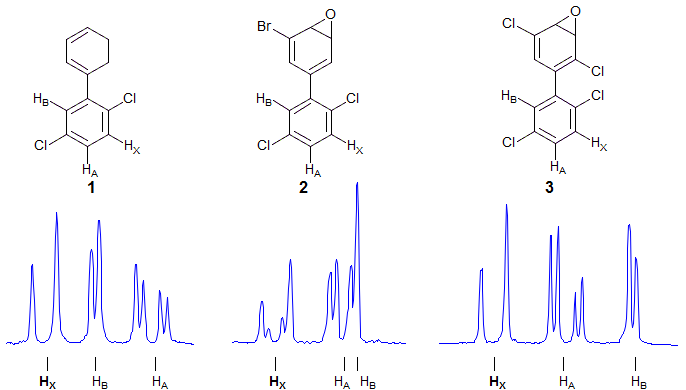
An important generalization in this area is that the severity of the "virtual coupling" effects is very strongly dependent on JAB - if JAB is larger than JAX and JBX then the X part is profoundly changed (i.e. doublet to triplet). On the other hand, when JAB is smaller than JAX or JBX then the perturbations are much less dramatic, leading to small additional lines and minor errors in apparent coupling constants. A case of this type is provided by the three compounds below. The aromatic protons are well separated in 1 and 3 and thus are pretty much first order, but in 2 HA appears nearly on top of HB. As a result, HX shows non-first order structure.
One of the main reasons that complex NMR spectra are simpler to interpret at higher field strengths is that many virtual coupling effects are ameliorated or disappear altogether as chemical shift differences (in Hz) become larger. Exceptions are AA'BB', AA'XX' patterns and more complicated analogs (such as AA'BB'X). These are always non-first order (if there is coupling between A and A' or B and B') because they always satisfy the criteria for virtual coupling: A and A' have zero chemical shift at any field strength. If A and A' are also coupled to each other (JAA' > 0) then the A and B protons, or any other protons coupled to A or B can give complex or misleading multiplets.
What are those Impurities? A student in the course brought me the NMR spectrum of acrolein shown below, with the question: why can't I get rid of the impurities, I've distilled the compound twice?
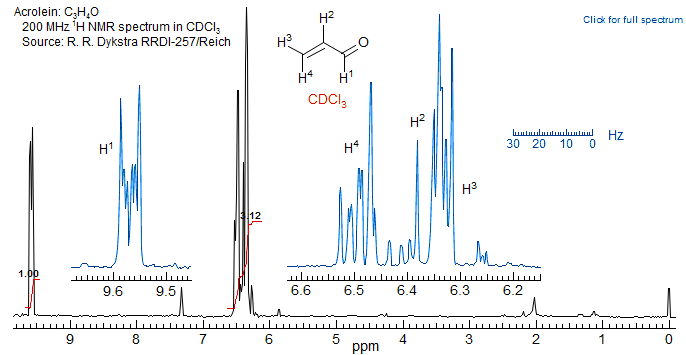
The answer? The compound is perfectly pure, as shown by its NMR spectrum in benzene-d6, which is essentially first order. The problem with the spectrum in CDCl3 is that two of the protons (H2 and H3) are nearly superimposed and are strongly coupled, so that all the others which are coupled to them become very complicated. Benzene, like other aromatic solvents, causes significant upfield shifts (especially in molecules with a high dipole moment, like this one) which removes the degeneracy and leads to an essentially first-order spectrum.
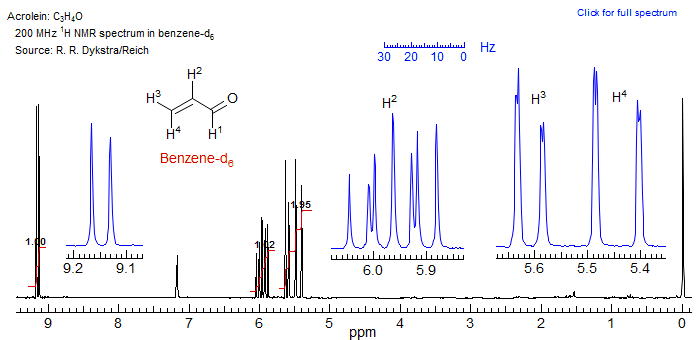
Exercise: Use WINDNMR to simulate first the benzene-d6 spectrum, and then the CDCl3 spectrum (all you should need to do is adjust the chemical shifts). Which two protons are superimposed in the CDCl3 spectrum?
Is it the Right Stuff? Accidental coincidences of chemical shifts and the resulting virtual coupling effects can lead to surprisingly deceptive NMR spectra, sufficiently so that the unwary researcher could conclude that a synthesis had failed. An interesting case is the phenyl region of the NMR spectrum below:
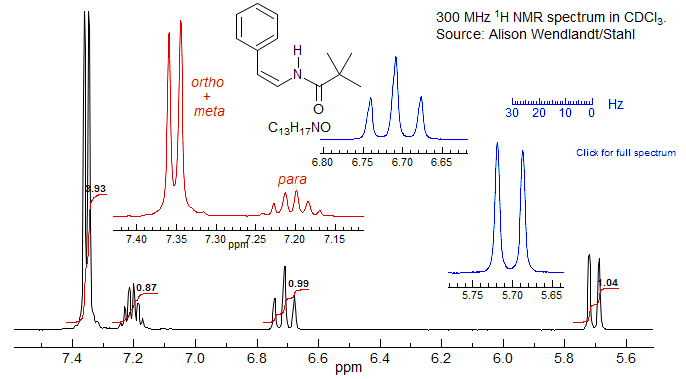
Taken at face value the aromatic region of the cis isomer looks like a 4H doublet and a 1H multiplet (pentet or sextet) with J of 4.3 Hz, hardly compatible with a monosubstituted phenyl group. Yet this is simply a phenyl group in which the ortho and meta protons accidentally have identical chemical shifts (or nearly so). Since they are strongly coupled to each other, the four protons behave as a unit, and the para proton appears to be equally coupled to all four, leading to the apparent doublet and pentet - a classical case of virtual coupling. The apparent coupling constant of 4.3 Hz is the average of the ortho and meta couplings.
In the trans isomer below the meta and para protons are a little further apart, and easily recognized for what they are (although still not even close to a first order pattern). For other similar examples see: 1, 2.
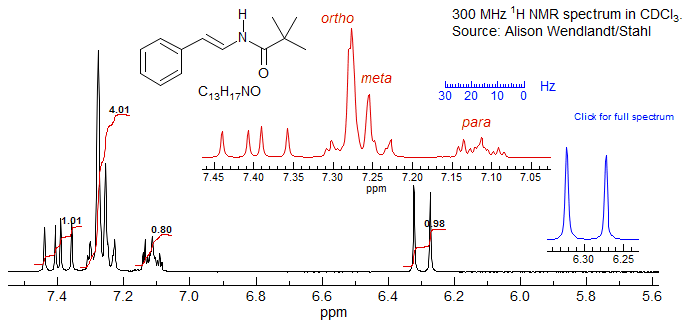
Why is my Spectrum so Ugly? In this spectrum there is an accidental superposition of two protons, HM and HN, which are coupled to each other. All of the protons coupled to these two, HA, HB, HX, and HY, show strongly second order distorted multiplets, with extra lines and a non-centrosymmetric structure. A simple method to address this kind of problem is to take the spectrum in an aromatic solvent (or even just add a few drops of benzene-d6 or pyridine to the sample), which in many cases moves the protons around enough that the second-order effects are reduced or eliminated. When the spectrum is taken in benzene, HM and HN are shifted away from each other, and HA and HB now show more or less first order multiplets.
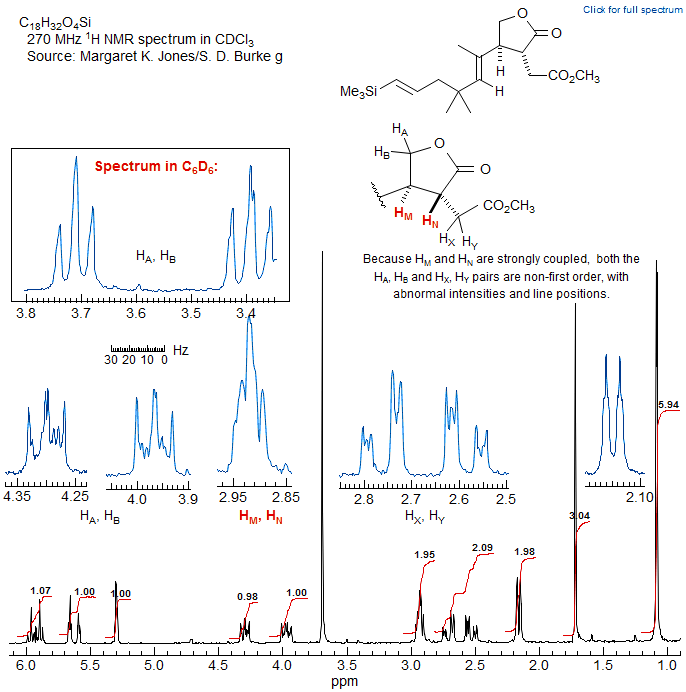
Methyl Region AMX3 ---> ABX3 ---> AA'X3
Methyl groups usually give reliably simple multiplets in NMR spectra. However, if a methyl group is coupled to a proton which is part of a strongly-coupled system, then its NMR signal can be complex and give misleading information. For examples: 1.
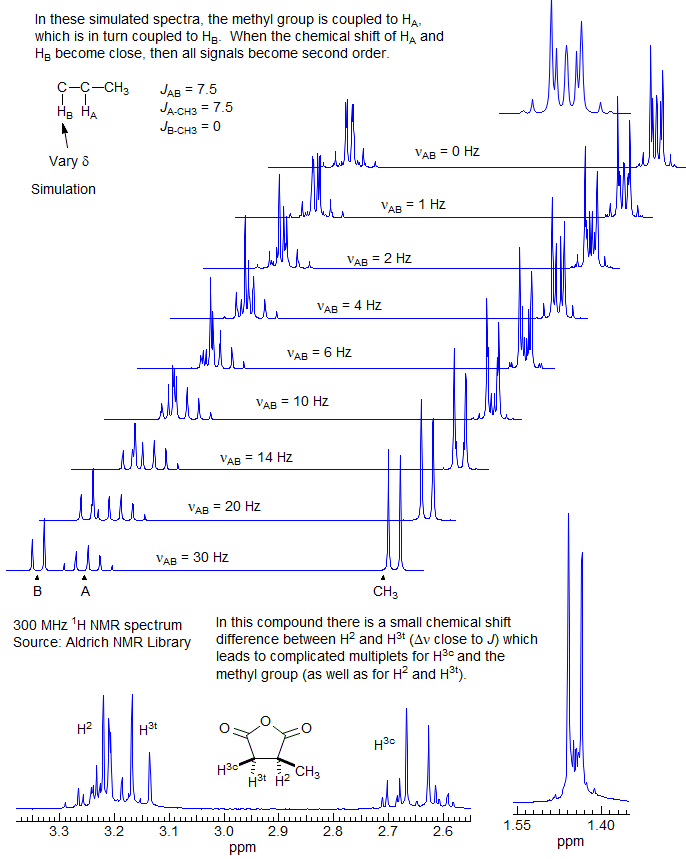
The kind of shift coincidences that cause strong second-order effects are especially pronounced in systems where the chemical shifts are identical by symmetry, In the example below of 2-butene epoxide both the CH3 and the CH signals are complex:
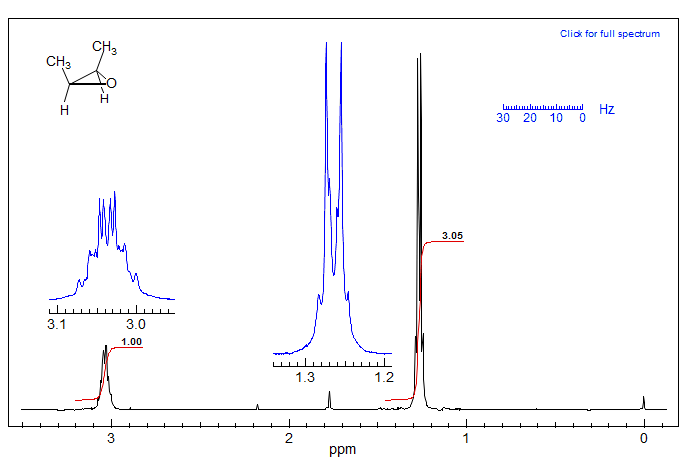
Approximate Nature of "AMX" Coupling ConstantsAn example of how virtual coupling effects can distort NMR coupling behavior is provided by an analysis of the multiplet at δ 1.6 in the spectrum below (protons M and N). The multiplet can be analyzed as a more or less first order pattern, giving the coupling constants shown.
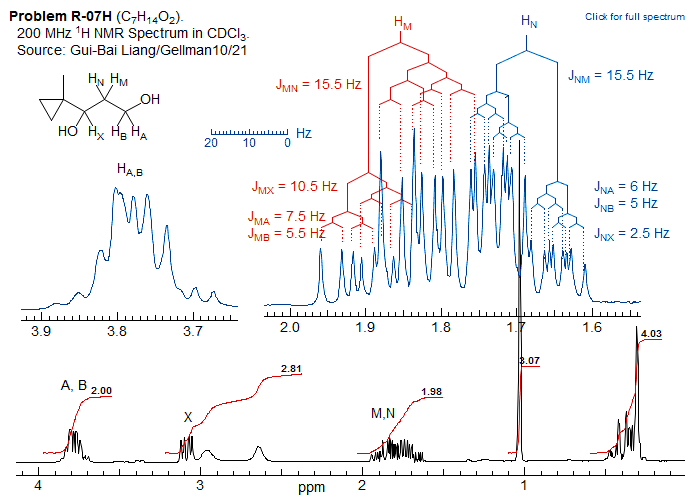
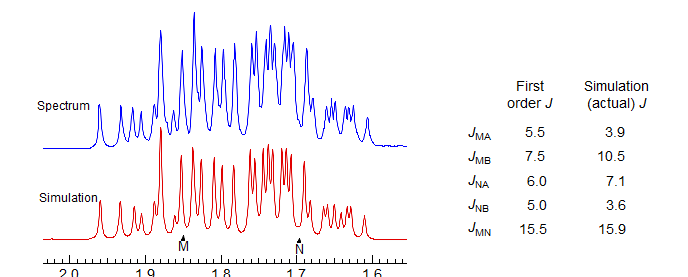
However, a proper simulation of this 5-spin system with WINDNMR leads to quite different coupling constants between the M,N and A,B protons. This is because the A and B protons (at δ 3.8) are nearly superimposed, so that the apparent coupling constants (e.g. 5.5 and 7.5) tend towards the average of the actual ones (3.9 and 10.5). Note that JMN is measured nearly correctly in the first order analysis: 15.5 Hz, vs 15.9 in the simulation.
Previous Section: AA'XX' and AA'BB' Patterns · Home